General Bioinformatics analysis: Gene and protein information (e.g., expression, location, function), Primer designing, RAPD analysis, Sequence analysis (e.g., BLAST, MSA, Phylogenetics), etc.
Sequencing: DNA, RNA and amplicon sequencing NGS sequencers (we work with partnering companies).
Creation of custom designed databases and software
Data mining and biocuration
Clinical data analysis
Advanced literature review and meta-analysis
Biomarker discovery by in–silico approaches
Developing databases and performing gene expression* analysis using microarray* and RNA sequencing data [multiple projects and collaborations; 3 of the collaborative work published]
Metagenomics* analysis of sequencing data from NGS & Sanger's method in the context of human gut microbiome [completed for a medical research group in India]
Reference and de novo genome sequence assembly*, gene annotations (miRNA included), phylogenetic analysis of raw genome sequence reads [multiple projects]
miRNA (microarray)* data analysis [completed for a research group at CCMB, Hyderabad, India]
Exon-array (Affymetrix)* data analysis for brain tissue [completed for a research group at CCMB, Hyderabad, India]
CHiP-seq* data analysis [completed for a research group at Kidwai Memorial Institute of Oncology, Bengaluru, India]
Primer and mutation* analysis in the context of oral cancer [completed for a company, Malaysia]
Protein interaction and pathway analysis applied to mass spec and microarray data in the context of a human disease [completed for a research group at NIRRH, Mumbai, India]
Meta analysis across cancer data sets for specific human genes of interest [completed for a research group at NIRRH, Mumbai, India]
Transcription regulatory network analysis of human genes of importance for reproduction in males [completed for a research group at NIRRH, Mumbai, India]
Development of a novel meta-analysis algorithm to make use of different types of gene expression data [in-house]
Development of a new computational platform for protein-interaction analysis [in-house]
Development of new RNA-protein interaction database by biocuration [in-house]
Development of online test system for screening candidates for course/job-suitability in the area of life sciences [in-house]
Development of a software for compiling hits obtained by literature search across multiple important resources [in-house]
Computational structural analysis to screen and dock ligands [collaboration with VIT, Tamilnadu, India; published]
*We can also carry out some of the initial wet-lab work (purifications, quality checks etc) or any other molecular biology laboratory work, in some cases, with the help of our collaborating companies.
Manu S, Acharya KK*, Thiyagarajan S (2018) Systematic analyses of autosomal recombination rates from the 1000 genomes project uncovers the global recombination landscape in humans. bioRxiv PrePrint.
Adurthi S, Kumar MM, Vinodkumar HS*, Mukherjee G, Krishnamurthy H, Acharya KK*, Bafna UD, Uma DK, Abhishekh B, Sudhir Krishna, Parchure A, Murali A, and Jayshree RS (2017) Oestrogen Receptor-α binds the FOXP3 promoter and modulates regulatory T-cell function in human cervical cancer. Scientific Reports 7 7(1):17289
Narayanaswamy PB, Baral TK*, Haller H, Dumler I, Acharya KK* and Kiyan Y (2017) Transcriptomic pathway analysis of urokinase receptor silenced breast cancer cells: a microarray study. Oncotarget 8(60): 101572-101590.
Kouser. K, P. G. Lavanya, Rangarajan L, Acharya KK* (2016) Effective Feature Selection for Classification of Promoter Sequences. PLOS One 11(12): e0167165.
Kumar MM, Davuluri* S, Poojar S, Mukherjee G, Bajpai AK*, Bafna UD, Devi UK, Kallur PP, Acharya KK*, Jayshree RS. (2016) Role of estrogen receptor alpha in human cervical cancer-associated fibroblasts: a transcriptomic study. Tumour Biol. Apr;37(4): 4409-20.
Pathak BR, Breed AA, Apte S, Acharya KK*, Mahale SD. Cysteine-rich secretory protein 3 plays a role in prostate cancer cell invasion and affects expression of PSA and ANXA1. Mol Cell Biochem. 2016; 411(1-2):11-21.
Chandrashekar DS, Dey P, Acharya KK* (2015) GREAM: A web server to short-list the potentially important genomic repeat elements based on over-/under-representation in specific chromosomal locations such as the gene neighborhoods within or across 17 mammalian species. PLOS One Jul 24;10(7):e0133647.
Roy D., Kumar V*, Acharya KK*, Thirumurugan K. Probing the Binding of Syzygium-Derived α-Glucosidase Inhibitors with N- and C-Terminal Human Maltase Glucoamylase by Docking and Molecular Dynamics Simulation. Applied biochemistry and biotechnology, [2014] 172(1):102-14
Bhagwat S, Dalvi V, Chandrasekhar DS, Matthew T, Acharya KK* Acetylated alpha tubulin is reduced in individuals with poor sperm motility. Fertility & Sterility,[2014] 101(1):95-104.e3
Chitturi N, Balagannavar G, Chandrashekar DS, Abinaya S*, Srini VS, Acharya KK*. TIPMaP: a web server to establish transcript isoform profiles from reliable microarray probes. BMC Genomics, [2013] 14: 922
Bhagwat SR, Chandrashekar DS, Kakar R, Davuluri S*, Bajpai AK*, Nayak S, Bhutada S, Acharya KK*, Sachdeva G. Endometrial receptivity: a revisit to functional genomics studies on human endometrium and creation of HGEx-ERdb. PLOS One, [2013] 8(3), e58419
Bajpai AK*, Davuluri S*, Chandrashekar DS., Ilakya S, Dinakaran M.* and Acharya KK*. MGEx-Udb: A Mammalian Uterus Database for Expression-Based Cataloguing of Genes across Conditions, Including Endometriosis and Cervical Cancer PLOS One, [2012], 7(5), e36776
Bajpai AK*, Davuluri S*, Haridas H, Kasliwal G, Deepti H, Sreelakshmi KS, Chandrashekar DS, Bora P, Farouk M, Chitturi N, Samudyata V, ArunNehru KP & Acharya KK*. In search of the right literature search engine(s) Nature Precedings, [2011]
Acharya KK*, Chandrashekar DS, Chitturi N, Shah H, Malhotra V, Sreelakshmi KS, Deepti H, Bajpai A*, Davuluri S*, Bora P, Rao L. A novel tissue-specific meta-analysis approach for gene-expression predictions, initiated with a mammalian gene expression testis database. BMC Genomics, [2010], 11: 467
Kshitish Acharya K*: Capacities vs. jobs in bioinformatics and biotechnology: a few points to the attention of current students & job-seekers [2014] In: "Biotech Career Ready Reckoner 2014" (BCRR 2014), Vivify Media Private Limited & Department of Biotechnology (DBT), Govt. of India.
*Researchers who had contributed from Shodhaka Life Sciences.








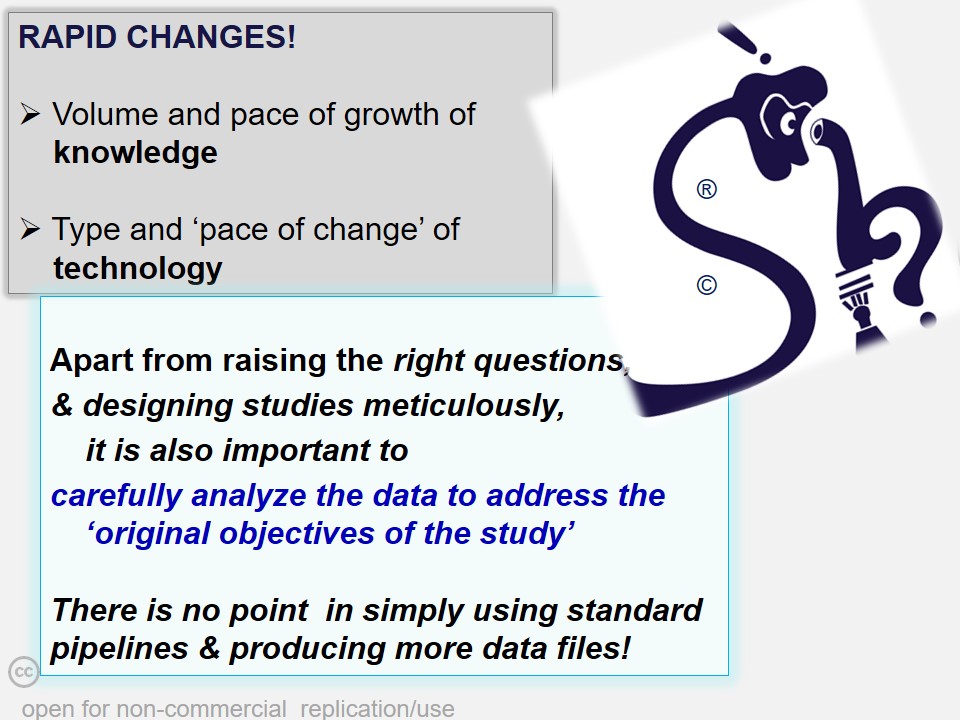
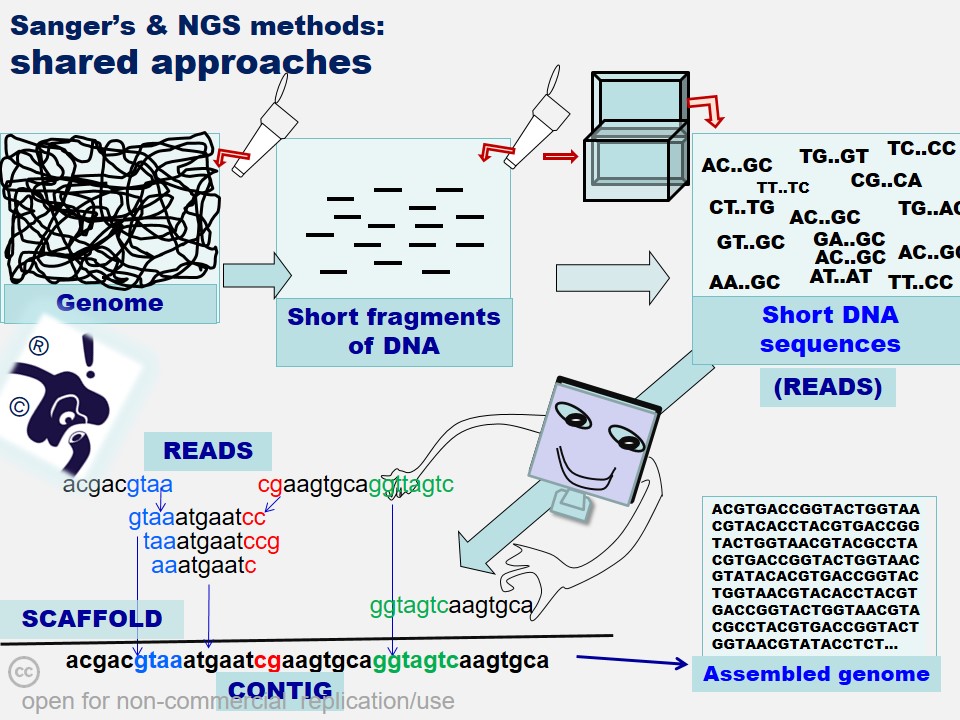


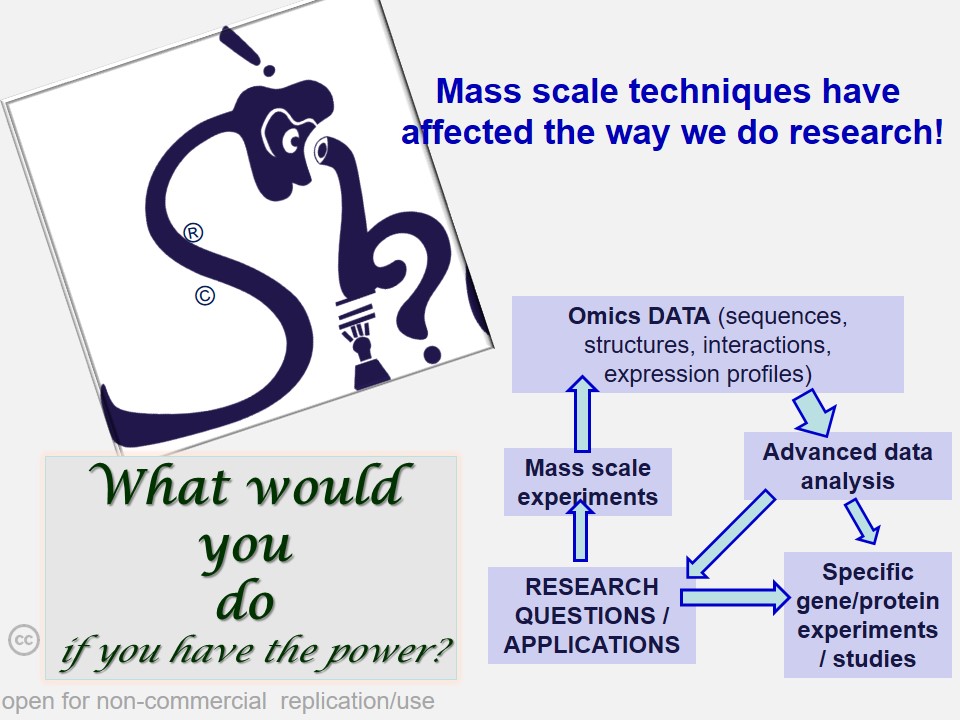
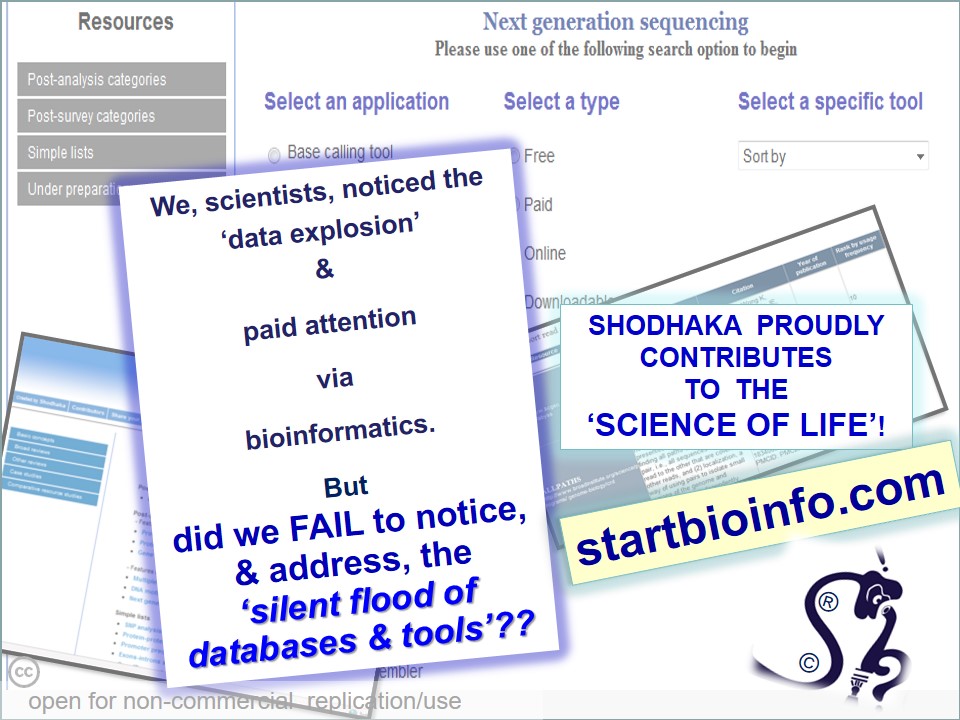
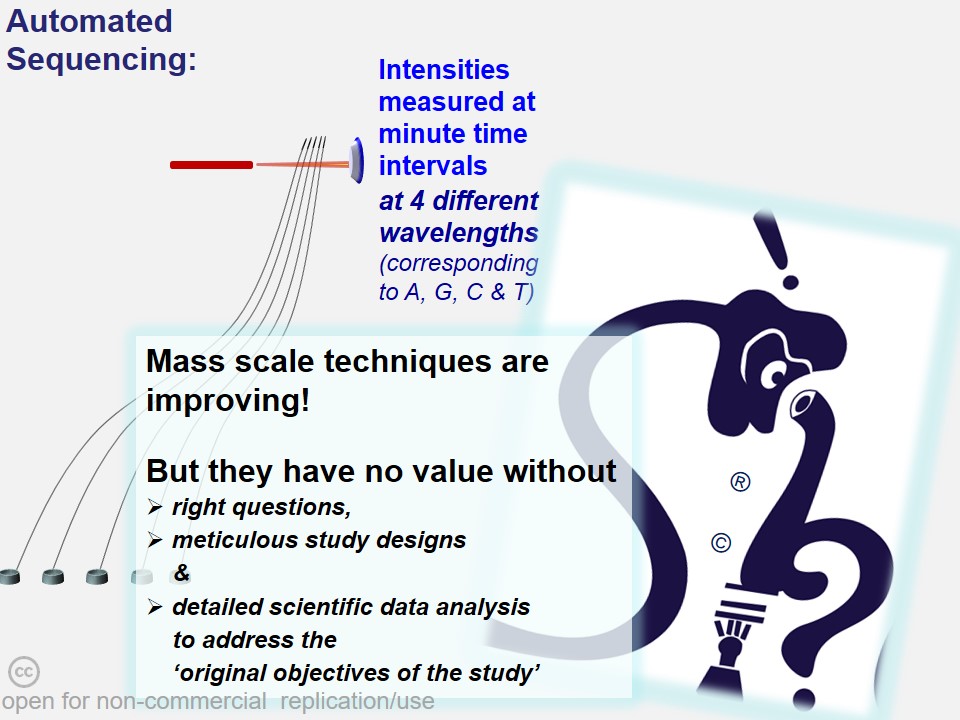
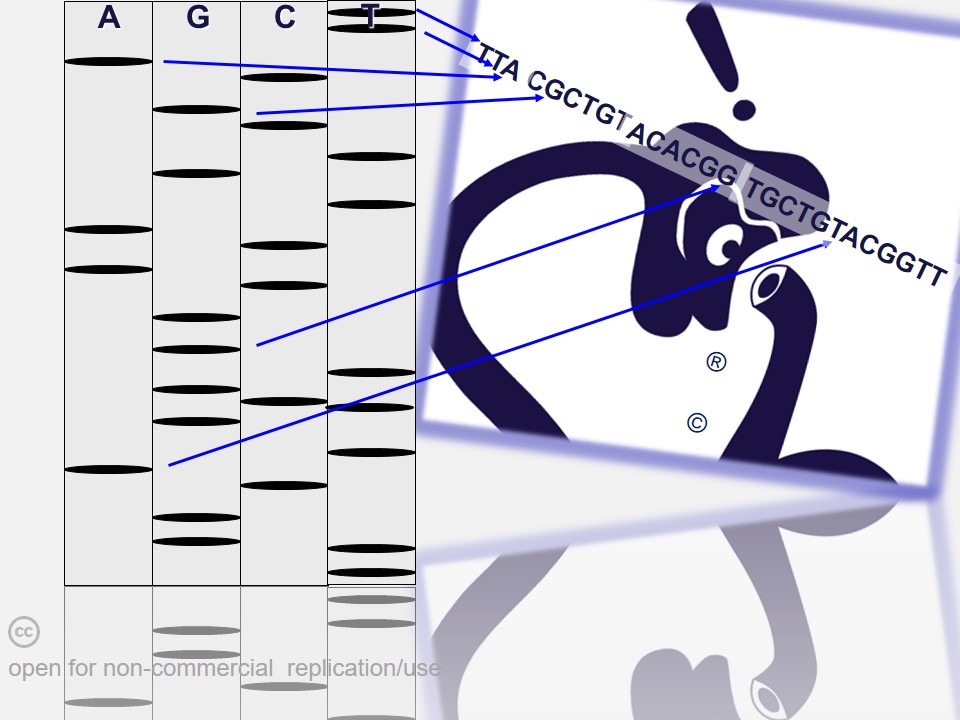

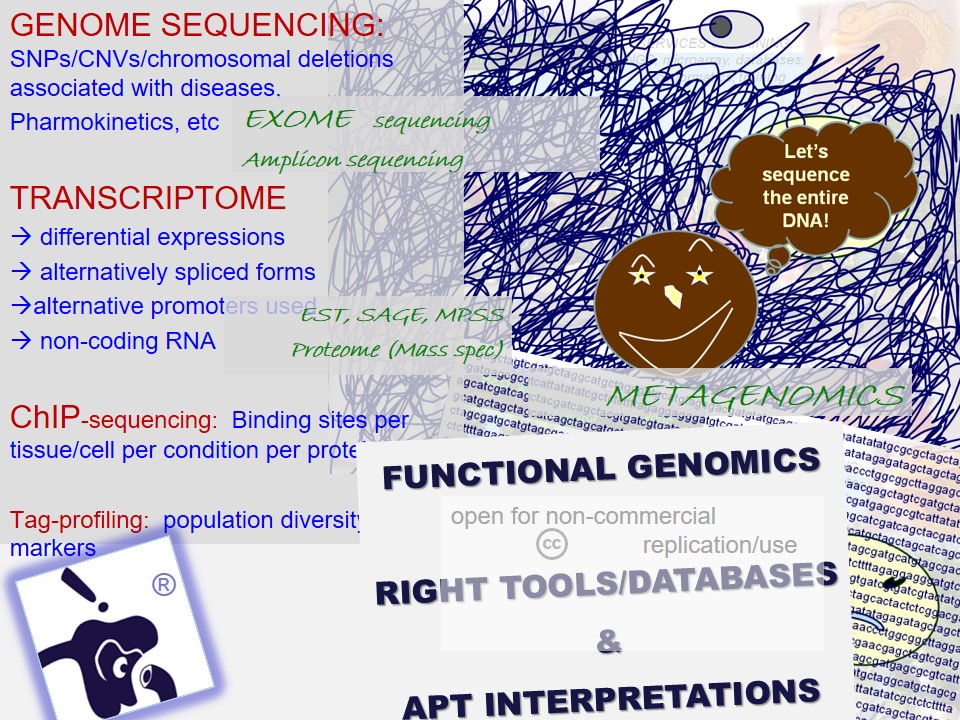
In many cases, critical data mining, data analysis and interpretations of scientific work suffers from a casual approach. 'Shodhaka' began with a vision to effectively address mainly two such imperfections in data mining and analysis: a) passive approach that is disconnected from the original objectives of the scientists; b) lack of objective selection and utilization of biological databases and software.
Established in 2009, the young team of scientists at 'Shodhaka' began with the herculean task of compiling, cataloguing and scientifically comparing the existing databases and software. The public web-portal startbioinfo.com - launched in 2010, and some of our publications illustrate our efforts in this direction. We are perhaps the only organization (at least a commercial one) that has spent more than 8 years in systematically comparing several types of such bioinformatic resources. This work has helped us to add value to services to the research projects of our client-scientists. The work not only helped us to determine the right choices among the tools and databases for specific tasks, but also identify gaps in the availability of such resources for certain applications. In such cases, we have been developing required databases and software for public use in collaboration with IBAB (www.ibab.ac.in), an institute that promotes entrepreneurship and where the founder director (Prof. Kshitish Acharya) continues to serve as a faculty scientist.
We have now completed multiple service-projects in the areas of NGS and microarray data analysis. We have also been active in the areas of biocuration, custom-designing databases & software, biomarker discovery by in silico approaches, and assisting scientists in manuscript writing/refining. We are proud that most client scientists are impressed with 'quality of work', and have particularly appreciated our tertiary data analysis contributions to their research. We have become the preferred service providers of most of the scientists who used our services. We hope to continue to do so. We have grown slowly but steadily without much of marketing efforts. We excited about the possibility reaching more researchers and contributing to their scientific endeavors. Do contact us please, if you have any bioinformatic or biostatistic need.


For current openings, please read the details below and send your CV as per instructions for current or possible future positions.
We are currently also looking for,
a) Individuals who are wish to be a 'marketing leader'. If you have a science background and confident of your communication abilities in English language - apply now. Experience is preferable, but not mandatory.
INSTRUCTIONS:
1. CV should be in word/pdf format
2. Name of the CV file should have the job applying for (e.g., writer, marketing, others) followed by your first or last name (e.g., Marketing_Archana)
and send to shodhaka.ls@gmail.com
How suitable are you across different type of jobs today in biotech-sector?
Shodhaka offers SOTS-JSA that can be your online career counselor.
This unique system may also be your door for job-opportunities in many other companies/research organizations.
We also conduct various TRAINING-PROGRAMS AND WORKSHOPS
Possible future positions at 'Shodhaka':
You can apply with CV to (shodhaka.ls@gmail.com). We will consider your CV for any current and future openings.
Do not forget SOTS JSA, it can be your guide and gateway to any job and career!
*Remuneration for all the positions listed above will be based on the performance in the selection process and the training required.